
HIGHER ORDER ANOVA WITH REPEATED MEASURES If you're unfamiliar with repeated measures analysis, I suggest you look at the Single Factor tutorial first. Two Way Designs With Repeated Measures on Both Factors We will use a data set from the MASS package called "Rabbit". The description on the help page reads as follows:
"Five rabbits were studied on two occasions, after treatment with saline
(control) and after treatment with the 5-HT_3 antagonist MDL 72222. After
each treatment ascending doses of phenylbiguanide were injected intravenously
at 10 minute intervals and the responses of mean blood pressure measured.
The goal was to test whether the cardiogenic chemoreflex elicited by
phenylbiguanide depends on the activation of 5-HT_3 receptors." [Note:
5-HT_3 is a serotonin receptor.]
We will treat Treatment and Dose as factors (throwing out valuable numerical
information--tsk) and, thus, we have a 2x6 factorial design with repeated
measures on both factors. The response is BPchange. The subject ID is Animal.
> data(Rabbit, package="MASS")
> summary(Rabbit)
BPchange Dose Run Treatment Animal
Min. : 0.50 Min. : 6.25 C1 : 6 Control:30 R1:12
1st Qu.: 1.65 1st Qu.: 12.50 C2 : 6 MDL :30 R2:12
Median : 4.75 Median : 37.50 C3 : 6 R3:12
Mean :11.22 Mean : 65.62 C4 : 6 R4:12
3rd Qu.:20.50 3rd Qu.:100.00 C5 : 6 R5:12
Max. :37.00 Max. :200.00 M1 : 6
(Other):24
> Rabbit$Dose = factor(Rabbit$Dose) # make sure R sees your factors as factors
> with(Rabbit, table(Treatment, Dose)) # check for balance
Dose
Treatment 6.25 12.5 25 50 100 200
Control 5 5 5 5 5 5
MDL 5 5 5 5 5 5
> ### a more thorough check would be table(Subject,Treatment,Dose) showing
> ### that each rabbit appears once in each of the treatment cells
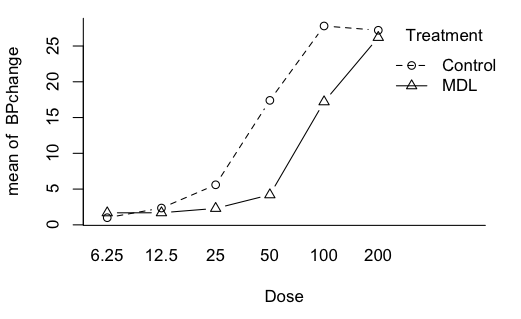
> with(Rabbit, tapply(BPchange, list(Treatment,Dose), mean))
6.25 12.5 25 50 100 200
Control 1.00 2.35 5.6 17.4 27.8 27.2
MDL 1.68 1.69 2.3 4.2 17.2 26.2
> with(Rabbit, tapply(BPchange, list(Treatment,Dose), var))
6.25 12.5 25 50 100 200
Control 0.15625 1.925 7.30 29.8 51.7 38.7
MDL 0.62325 0.538 1.45 8.7 62.2 47.7
We have some pretty ugly problems with the variances, but we're going to
have to ignore that, as data sets for balanced ANOVA problems are few and
far between in these built-in data sets. Let's see what effects we might have.
> with(Rabbit, interaction.plot(Dose, # factor displayed on x-axis
Treatment, # factor displayed as multiple lines
BPchange, # the response variable
type="b", # plot both points and lines
pch=1:2, # use point characters 1 and 2
bty="l")) # draw a box around the plot that looks like an L
> aov.out = aov(BPchange ~ Dose * Treatment + Error(Animal/(Dose*Treatment)), data=Rabbit)
> ### (Dose*Treatment) is the same as (Dose+Treatment+Dose:Treatment)
> summary(aov.out)
Error: Animal
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 4 351.4 87.86
Error: Animal:Dose
Df Sum Sq Mean Sq F value Pr(>F)
Dose 5 6022 1204.3 66.17 9.48e-12 ***
Residuals 20 364 18.2
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: Animal:Treatment
Df Sum Sq Mean Sq F value Pr(>F)
Treatment 1 328.5 328.5 13.65 0.0209 *
Residuals 4 96.3 24.1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: Animal:Dose:Treatment
Df Sum Sq Mean Sq F value Pr(>F)
Dose:Treatment 5 419.9 83.99 8.773 0.000156 ***
Residuals 20 191.5 9.57
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
There is a significant main effect of dose. No surprise there! There is a
significant main effect of Treatment, which is what we were supposedly
looking for. But the main effects are complicated by the presence of a
significant interaction.
Check the ANOVA table to make sure the error terms are right. Each treatment should have the corresponding subject x treatment interaction as its error term. The interaction of the treatments should have the subject x treatment x treatment interaction as its error term. Looks like we got that right! It's also a good idea to do a quick check of the degrees of freedom to make sure they are correct. One reason they might be wrong is if we failed to declare a variable coded numerically as a factor when we wanted to use it as such. Looks like we're good here, though. The extension to the three factor completely repeated measures design is straightforward. The error term would be Error(subject/(IV1*IV2*IV3)). Be sure you use the parentheses around the names of the IVs in the "denominator" of the error term. Otherwise, your Error will be in error. Two Way Mixed Factorial Designs We have to be careful with the term "mixed" when we're talking about experimental designs. It is used in two very different ways: mixed factorial design and mixed model. A mixed factorial design is one that has both between and within subjects factors. A mixed model is one that has both fixed and random effects. Here again we discover that the R people are not really ANOVA people.
We're going to have to make do by modifying a data set that is not ideally
suited to the problem. There is a data set called ChickWeight that I am going
to modify to make it a balanced mixed factorial. Follow these instructions
very carefully!
> aov.out = aov(weight ~ Diet * Time + Error(Chick/Time), data=CW)
> summary(aov.out)
Error: Chick
Df Sum Sq Mean Sq F value Pr(>F)
Diet 2 10946 5473 1.562 0.228
Residuals 27 94613 3504
Error: Chick:Time
Df Sum Sq Mean Sq F value Pr(>F)
Time 9 883951 98217 245.987 <2e-16 ***
Diet:Time 18 13236 735 1.842 0.0215 *
Residuals 243 97024 399
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
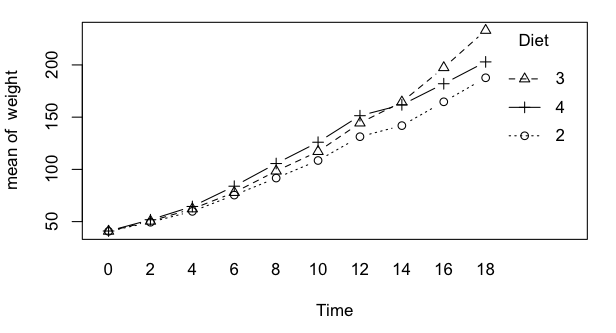
The correct error term for the between subjects factor is just subject
variability, as usual. The correct error term for the within subjects factor
is that factor crossed with subject. Same is true for the interaction. We are
two for two!
Well, not so fast! Let's think about this one for a minute. What problem
do you think we might have here? I'd bet my paycheck that the weight of
chicks is a whole lot less variable when they are 0 days old than it is when
they are 18 days old. I anticipate a serious problem with homogeneity of
variance, or rather the lack thereof.
Three Way Designs With Repeated Measures on One Factor We will use a built-in data set called CO2, which contains data
from an experiment on the cold tolerance of the grass species
Echinochloa crus-galli.
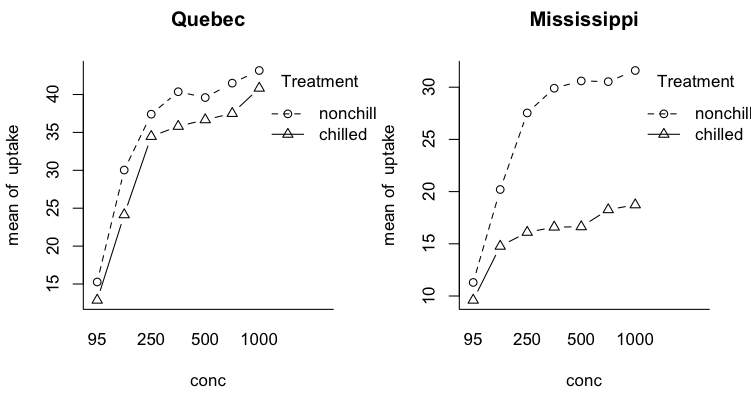
> aov.out = aov(uptake ~ Type * Treatment * conc + Error(Plant/conc), data=CO2)
> summary(aov.out)
Error: Plant
Df Sum Sq Mean Sq F value Pr(>F)
Type 1 3366 3366 95.195 1.02e-05 ***
Treatment 1 988 988 27.949 0.00074 ***
Type:Treatment 1 226 226 6.385 0.03543 *
Residuals 8 283 35
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: Plant:conc
Df Sum Sq Mean Sq F value Pr(>F)
conc 6 4069 678.1 172.562 < 2e-16 ***
Type:conc 6 374 62.4 15.880 5.98e-10 ***
Treatment:conc 6 101 16.8 4.283 0.001557 **
Type:Treatment:conc 6 112 18.7 4.748 0.000717 ***
Residuals 48 189 3.9
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Three Way Designs With Repeated Measures on Two Factors We have finally crapped out on built-in data sets. So we are going to have
to import one. If you have an Internet connection, you can try this, typing
with excruciating care. (If you don't have an Internet connection, then how
are you reading these tutorials?!)
The experiment was similar to studies done by Donders in the 19th century
in which he was attempting to measure the amount of time it takes a mental
event to occur (mental chronometry it was called). Simple reaction time tasks
are quite simple. You see a stimulus, and you react to it as quickly as you
possibly can. Disjunctive tasks are a little more complex, in that two (or
more, but two here) different stimuli might be presented, and you are only to
respond to one of them. So in addition to the time it takes to react, there is
also a decision to make: should I react or not. Finally, in choice reaction
time tasks, there are two (or more) different stimuli, and each requires a
different response. So the decision has become more complex: which response
should I make.
> aov.out = aov(RT ~ lab * task * color + Error(subject/(task*color)), data=react)
> summary(aov.out)
Error: subject
Df Sum Sq Mean Sq F value Pr(>F)
lab 1 26289 26289 8.756 0.00923 **
Residuals 16 48039 3002
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: subject:task
Df Sum Sq Mean Sq F value Pr(>F)
task 2 106509 53255 23.591 5.06e-07 ***
lab:task 2 9448 4724 2.093 0.14
Residuals 32 72238 2257
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Error: subject:color
Df Sum Sq Mean Sq F value Pr(>F)
color 1 290 290.1 0.260 0.617
lab:color 1 169 168.7 0.151 0.702
Residuals 16 17831 1114.4
Error: subject:task:color
Df Sum Sq Mean Sq F value Pr(>F)
task:color 2 61 30.6 0.020 0.980
lab:task:color 2 4366 2182.8 1.428 0.255
Residuals 32 48899 1528.1
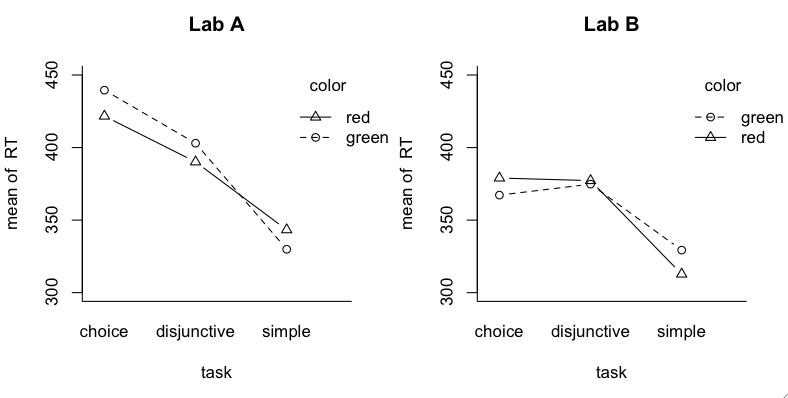
There are two significant main effects, lab and task. Task I expected. Lab I
have no explanation for considering the very simple nature of the response.
A Final Note I don't know how to do these problems in R using a multivariate approach. If anyone does, please clue me in. created 2016 February 9 |