
| Table of Contents
| Function Reference
| Function Finder
| R Project |
MAKING R WORK FOR YOU
In the course of working through these tutorials, you may have noticed that,
just in places, R is a twinkling shy of what might be referred to as
perfection. This occurs mostly in three ways: functions with arcane names,
functions with goofy syntax, and functions that don't exist, such as ones for
sum of squares and standard error of the mean. In this tutorial we're going
to take care of a lot of that.
My intention is to someday bundle together a lot of what is done in this
tutorial and offer it as an optional package that can be downloaded from
CRAN. Someday. In the meantime, I'm going to show how to create your own bundle
of custom functions, which can be continually modified, and which you can use
without cluttering up your workspace with custom functions.
Before you read this tutorial, you should read
Writing Your Own Functions and Scripts.
> setwd("Rspace") # very important (reasonably important anyway!)
> rm(list=ls()) # a clean workspace is essential
Optional Packages
Before we get to writing our own stuff, it is worthwhile to have one of the
optional packages available at CRAN. Frankly, I think this should be part of
the default installation. One Note: When I installed it, I got a warning about
version numbers. I installed it anyway and haven't had a problem, but you might
want to consider upgrading to the latest version of R first.
> install.packages("car") # see the Package Management tutorial
When you do this, R will open a list of mirror sites for you, from which you
should choose one near you. Once you do that, the rest happens automatically.
Let's see what we're getting.
> library(car) # attach to the search path in position 2
> ls(pos=2) # see the contents
[1] "Adler" "AMSsurvey" "Angell"
[4] "Anova" "Anscombe" "av.plot"
[7] "av.plots" "avPlot" "avPlots"
[10] "basicPower" "basicPowerAxis" "Baumann"
[13] "bc" "bcPower" "bcPowerAxis"
[16] "Bfox" "Blackmore" "Boot"
[19] "bootCase" "box.cox" "box.cox.powers"
[22] "box.cox.var" "box.tidwell" "boxCox"
[25] "boxCoxVariable" "Boxplot" "boxTidwell"
[28] "Burt" "CanPop" "carWeb"
[31] "ceres.plot" "ceres.plots" "ceresPlot"
[34] "ceresPlots" "Chile" "Chirot"
[37] "compareCoefs" "confidence.ellipse" "confidenceEllipse"
[40] "contr.Helmert" "contr.Sum" "contr.Treatment"
[43] "cookd" "Cowles" "cr.plot"
[46] "cr.plots" "crp" "crPlot"
[49] "crPlots" "data.ellipse" "dataEllipse"
[52] "Davis" "DavisThin" "deltaMethod"
[55] "densityPlot" "Depredations" "dfbetaPlots"
[58] "dfbetasPlots" "Duncan" "durbin.watson"
[61] "durbinWatsonTest" "dwt" "ellipse"
[64] "Ericksen" "estimateTransform" "Florida"
[67] "Freedman" "Friendly" "gamLine"
[70] "Ginzberg" "Greene" "Guyer"
[73] "Hartnagel" "hccm" "Highway1"
[76] "Identify3d" "infIndexPlot" "influenceIndexPlot"
[79] "influencePlot" "inverseResponsePlot" "invResPlot"
[82] "invTranEstimate" "invTranPlot" "KosteckiDillon"
[85] "Leinhardt" "levene.test" "leveneTest"
[88] "leverage.plot" "leverage.plots" "leveragePlot"
[91] "leveragePlots" "lht" "linear.hypothesis"
[94] "linearHypothesis" "linearHypothesis.default" "loessLine"
[97] "logit" "makeHypothesis" "Mandel"
[100] "Manova" "marginalModelPlot" "marginalModelPlots"
[103] "matchCoefs" "mcPlot" "mcPlots"
[106] "Migration" "mmp" "mmps"
[109] "Moore" "Mroz" "ncv.test"
[112] "ncvTest" "nextBoot" "OBrienKaiser"
[115] "Ornstein" "outlier.test" "outlierTest"
[118] "panel.car" "Pottery" "powerTransform"
[121] "Prestige" "printHypothesis" "probabilityAxis"
[124] "qq.plot" "qqp" "qqPlot"
[127] "quantregLine" "Quartet" "recode"
[130] "Recode" "regLine" "residualPlot"
[133] "residualPlots" "Robey" "Sahlins"
[136] "Salaries" "scatter3d" "scatterplot"
[139] "scatterplot.matrix" "scatterplotMatrix" "showLabels"
[142] "sigmaHat" "SLID" "slp"
[145] "Soils" "some" "sp"
[148] "spm" "spread.level.plot" "spreadLevelPlot"
[151] "States" "subsets" "symbox"
[154] "testTransform" "Transact" "UN"
[157] "USPop" "vif" "Vocab"
[160] "wcrossprod" "WeightLoss" "which.names"
[163] "whichNames" "Womenlf" "Wong"
[166] "Wool" "yjPower" "yjPowerAxis"
That is a lot of stuff! Many of those items are data sets, such as Adler, for
example. Help pages are available if you want details. There are also a few
functions here that appear to be available in the base packages, for
example, Anova and Boxplot. The function name is capitalized, however, so as
not to mask the similar function in base R. The "car" versions work somewhat
differently.
Just a few things here worth pointing out are Anova, which allows Type II
and Type III tests for unbalanced factorial designs (Type II is default, in
fact), leveneTest, which computes a Levene's test for homogeneity of variance,
Recode and recode, which allow more convenient recoding of a numeric vector
than is available in base R, and vif, which calculates variance inflation
factors for linear models. In fact, there is so much good stuff here that I
will just have to leave you to investigate it for yourself. This package was
developed to accompany the very excellent R manual "An R Companion to Applied
Regression" (2nd ed.) by Fox and Weisberg (2011), which I can highly recommend
for an introduction to much of what is in "car".
> detach("package:car")
I'm not suggesting that this is the only valuable optional package at CRAN,
and may not even be the most valuable one to you, but I have found it to be
almost indispensable.
A Few Simple One-Liner Custom Functions That Work on Vectors
To begin, make sure your workspace is clean, and I mean spotless! I would
also recommend if you have a .RData file in your current working directory
that you do not want to lose, you should load it and rename it. Then once
again, clean out your workspace. Your workspace should be character(0)!
You may recall that the length() function has
a shortcoming. It counts missing values, and there is nothing you can do to
stop it. Thus, if you want to know how many nonmissing values there are in a
vector--how many values actually went into a calculation--the length() function may not be your best bet. Here is a simple
replacement. The workings of this function was explained in a previous tutorial.
> sampsize = function(X) length(X) - sum(is.na(X))
And test it. (NOTE: I don't mean these tests to be rigorous, but these
functions should work. Here I just want to show that they give an answer. If
anyone sees a fault in any of these functions, please let me know.)
> ls()
[1] "sampsize"
> length(rivers)
[1] 141
> sampsize(rivers)
[1] 141
> length(islands)
[1] 48
> sampsize(islands)
[1] 48
> rivers[c(10:15)] = NA # won't hurt the permanent version
> length(rivers)
[1] 141
> sampsize(rivers)
[1] 135
>rm(rivers)
There is no sum of squares function in R. This is not too much of an oversight,
because you rarely would need such a function in most statistical work. But you
sure need one when you're first learning statistics, AND when you're trying to
teach it!
> SS = function(X) sum(X^2, na.rm=TRUE) - sum(X, na.rm=TRUE)^2 / (length(X) - sum(is.na(X)))
This should work on any numeric vector, even one with missing values.
> ls()
[1] "sampsize" "SS"
> SS(rivers)
[1] 34147177
> rivers[10:20] = NA
> SS(rivers)
[1] 33177355
> rm(rivers)
> SS(c(1,2,3)) # because we know the answer to this one!
[1] 2
> SS(c(1,2,NA))
[1] 0.5
Likewise, a function for standard error of the mean.
> sem = function(X) sqrt(var(X, na.rm=TRUE) / (length(X) - sum(is.na(X))))
And test it.
> ls()
[1] "sampsize" "sem" "SS"
> sqrt(var(rivers)/length(rivers))
[1] 41.59143
> sem(rivers)
[1] 41.59143
> rivers[10:20] = NA
> sem(rivers)
[1] 44.47893
> rm(rivers)
> sem(c(1,2,3))
[1] 0.5773503
> sqrt(1/3)
[1] 0.5773503
It annoys me that the name and syntax of the tapply()
function are so goofy. So I'm going to write a simple one-line replacement. (The
default argument labels in tapply are X, INDEX, and FUN, and my students never
get it).
> bygroups = function(data, groups, FUN=mean) tapply(data, groups, FUN)
This should do the means by default but can also be used with other functions.
> ls()
[1] "bygroups" "sampsize" "sem" "SS"
> head(chickwts)
weight feed
1 179 horsebean
2 160 horsebean
3 136 horsebean
4 227 horsebean
5 217 horsebean
6 168 horsebean
> with(chickwts, tapply(weight, feed, mean))
casein horsebean linseed meatmeal soybean sunflower
323.5833 160.2000 218.7500 276.9091 246.4286 328.9167
> with(chickwts, bygroups(data=weight, groups=feed))
casein horsebean linseed meatmeal soybean sunflower
323.5833 160.2000 218.7500 276.9091 246.4286 328.9167
> with(chickwts, tapply(weight, feed, sd))
casein horsebean linseed meatmeal soybean sunflower
64.43384 38.62584 52.23570 64.90062 54.12907 48.83638
> with(chickwts, bygroups(data=weight, groups=feed, FUN=sd))
casein horsebean linseed meatmeal soybean sunflower
64.43384 38.62584 52.23570 64.90062 54.12907 48.83638
Finally, when I summarize a numeric variable, more often than not I want a
mean, a standard deviation, and a sample size. The
summary() function gives me only one of those things.
> summarize = function(X) c(mean=mean(X, na.rm=TRUE), SD=sd(X, na.rm=TRUE), N=sampsize(X))
And test it. Notice that I took a shortcut here. This function depends upon
the presence of the sampsize() function.
> ls()
[1] "bygroups" "sampsize" "sem" "SS" "summarize"
> mean(rivers)
[1] 591.1844
> sd(rivers)
[1] 493.8708
> length(rivers)
[1] 141
> summarize(rivers)
mean SD N
591.1844 493.8708 141.0000
> rivers[10:20] = NA
> summarize(rivers)
mean SD N
599.2231 507.1378 130.0000
> rm(rivers)
Now check your workspace to make sure it has NOTHING in it but these five
functions. Then save and clear your workspace.
> ls()
[1] "bygroups" "sampsize" "sem" "SS" "summarize"
> getwd() # just checking!
[1] "/Users/billking/Rspace"
> save.image("myfunctions.RData")
> rm(list=ls())
> ls()
character(0)
I will show you how these functions will be loaded and used later. For now,
on to more complex stuff!
Making a Modification or Two
I'm not entirely happy with our sampsize()
function. First of all, I don't like the name. I also don't like that, as the
length() function does, it does things its way and
offers us no opportunity to modify that. What I want is a length function with
an na.rm option.
Open a script window (File > New Script in Windows or File > New Document on
a Mac), and type this.
size = function(X, na.rm=TRUE)
{
ifelse(na.rm,
length(X) - sum(is.na(X)),
length(X))
}
Then choose Edit > Run All in Windows, or highlight all lines of the script
and choose Edit > Execute on a Mac. This should put a function in your
workspace called size(). The function first tests
na.rm. If it's true, the second line is executed, and the function works like
sampsize. If it's false, then the third line is executed, and the function
works like length. The default is true. If you want false, you have to set the
option when you use the function.
> size(rivers)
[1] 141
> size(1:100)
[1] 100
> size(c(1:98,NA,NA))
[1] 98
> length(c(1:98,NA,NA))
[1] 100
> size(c(1:98,NA,NA), na.rm=F)
[1] 100
Check your workspace. The new function should be the ONLY thing there. If not,
make it so. Then...
> load("myfunctions.RData")
> rm(sampsize)
> save.image("myfunctions.RData")
...and your myfunctions.RData file is modified. What's the problem? Our
summarize() function depends upon the presence of
sampsize, and we've just undone that. It was kind of a questionable thing to
do in the first place, so we're going to fix it by amending the
summarize() function. Once again in the script window (delete anything
that is still there)...
summarize = function(X)
{
c(MEAN=mean(X, na.rm=T),
MEDIAN=median(X, na.rm=T),
SD=sd(X, na.rm=T),
N=length(X)-sum(is.na(X)),
NAs=sum(is.na(X)))
}
Run/Execute it. Then test it.
> summarize(rivers)
MEAN MEDIAN SD N NAs
591.1844 425.0000 493.8708 141.0000 0.0000
> summarize(c(1:100))
MEAN MEDIAN SD N NAs
50.50000 50.50000 29.01149 100.00000 0.00000
> summarize(c(1:98,NA,NA))
MEAN MEDIAN SD N NAs
49.50000 49.50000 28.43413 98.00000 2.00000
> summarize(c(1:98,NA,NA,1000))
MEAN MEDIAN SD N NAs
59.10101 50.00000 99.62936 99.00000 2.00000
Looks like it works, and when we created it, it overwrote the old version that
was in the workspace. Now we need to save the workspace again to update the
myfunctions file. Then clear it and on to the next section with thee!
> save.image("myfunctions.RData") # good idea to ls() it first!
> rm(list=ls())
Error Bars, Critical r, and Cohen's d
In this and future sections of this tutorial, I am going to be typing my
functions into a script window. That way they will be much easier to fix if
(when!) I screw them up. Reminder: The script window is reached in Windows via
File > New Script, and on a Mac via File > New Document.
There is no facility in R for plotting error bars on bar graphs (or any
other kind of graph). Putting on my R apologist hat, I would say fine, because
bar graphs are for frequencies, not for means! But not everyone sees it that
way, and I must respect their right to be mistaken, so I'm told. So here is an
error.bars() function. If you want to just copy and
paste it into R to create the function, I'm okay with that.
error.bars = function (x, y, e, cbl = 0.03)
{
num.groups = length(x)
for (i in 1:num.groups) {
arrows(x[i], y[i] - e[i], x[i], y[i] + e[i], angle = 90,
length = cbl, code = 3)
}
}
The function must be sent three vectors: x = a vector of x-coordinates where
you want error bars plotted, y = a vector of y-coordinates where you want
error bars centered (same order as the x-coords), and e = a vector of errors
(usually standard errors) that give half the length of the error bars.
Bar graphs are tricky in R, because they don't plot bars where you might
expect to find them. Let me show you.
> summary(chickwts) # some data to demonstrate
weight feed
Min. :108.0 casein :12
1st Qu.:204.5 horsebean:10
Median :258.0 linseed :12
Mean :261.3 meatmeal :11
3rd Qu.:323.5 soybean :14
Max. :423.0 sunflower:12
> means = with(chickwts, tapply(weight, feed, mean)) # get and save means
> barplot(means)
> barplot(means, ylim=c(0,500)) # make room for error bars
> sds = with(chickwts, tapply(weight,feed, sd)) # get and save SDs
> lengths = with(chickwts, tapply(weight, feed, length)) # get and save Ns
> sems = sds / sqrt(lengths) # get sems
> barplot(means, ylim=c(0,500)) -> bp # I always forget this!
> bp # this is where the bars are plotted!
[,1]
[1,] 0.7
[2,] 1.9
[3,] 3.1
[4,] 4.3
[5,] 5.5
[6,] 6.7
> error.bars(x=bp, y=means, e=sems, cbl=.1) # draw the error bars
Save the output of your barplot command. That will store the x-coords of the
bars on the bar graph. The cbl=.1 option controls the length of the crossbar
on the error bars (default value .03). Now we've made a mess, and we must
clean it up!
> ls()
[1] "bp" "error.bars" "lengths" "means" "sds" "sems"
> rm(bp, lengths, means, sds, sems)
> ls()
[1] "error.bars"
R does a good job of calculating correlations and correlation matrices, but
it gives no indication of when these values are statistically significant. I
could always look it up in a table, but hey! What do I have a computer for?
Here is a simple function for r-critcal based on the relationship between r
and student's t.
r.crit = function (df, alpha = 0.05)
{
t1 = qt(p = alpha, df = df, lower.tail = FALSE)
r1 = sqrt(t1^2/(t1^2 + df))
t2 = qt(p = alpha/2, df = df, lower.tail = FALSE)
r2 = sqrt(t2^2/(t2^2 + df))
output = c(df, alpha, r1, r2)
names(output) = c("df", "alpha", "1-tail", "2-tail")
output = round(output, 4)
output
}
And here's an example of how it is used.
> cor(USArrests)
Murder Assault UrbanPop Rape
Murder 1.00000000 0.8018733 0.06957262 0.5635788
Assault 0.80187331 1.0000000 0.25887170 0.6652412
UrbanPop 0.06957262 0.2588717 1.00000000 0.4113412
Rape 0.56357883 0.6652412 0.41134124 1.0000000
> dim(USArrests)
[1] 50 4
> r.crit(df=48, alpha=.01) # default alpha is .05
df alpha 1-tail 2-tail
48.0000 0.0100 0.3281 0.3610
Finally, Cohen's d, there is no R function for Cohen's d. We certainly can't
have that!
Cohens.d = function(group1, group2)
{
group1 = group1[!is.na(group1)] # strip NAs from group 1
group2 = group2[!is.na(group2)] # strip NAs from group 2
SSE = var(group1)*(length(group1)-1)+var(group2)*(length(group2)-1)
dfE = length(group1)+length(group2)-2
MSE = SSE/dfE
pooled.s = sqrt(MSE)
diff = abs(mean(group1)-mean(group2))
t = diff/(pooled.s*sqrt(1/length(group1)+1/length(group2)))
c(diff=diff,t=t,Cohens.d=abs(mean(group1)-mean(group2))/pooled.s)
}
Send it two vectors that you have used or are about to use as the two groups
to be compared in a t-test.
> casein = chickwts$weight[chickwts$feed=="casein"]
> horsebean = chickwts$weight[chickwts$feed=="horsebean"]
> Cohens.d(horsebean, casein)
diff t Cohens.d
163.383333 7.019741 3.005674
And now we've got some cleaning up to do! Following that, we're going to
(carefully!) add these functions to the ones we programmed above, and then
we're going to save the whole bunch of them.
> ls()
[1] "casein" "Cohens.d" "error.bars" "horsebean" "r.crit"
> rm(casein, horsebean)
> ls() # can't be too safe!
[1] "Cohens.d" "error.bars" "r.crit"
> load("myfunctions.RData") # add the previous ones
> ls() # really! you can't be too safe!
[1] "bygroups" "Cohens.d" "error.bars" "r.crit" "sampsize" "sem"
[7] "SS" "summarize"
> save.image("myfunctions.RData") # save all of them
> rm(list=ls())
A note: You caught me! Notice the workspace listings above contain sampsize
and not size. Why's that? Because the 3rd section of this tutorial is the last
section I wrote, so when I was writing this and future sections of this tutorial,
it was still sampsize, size having not yet been created. Now on to even more
complex stuff!
Some Functions Related to ANOVA
With apologies to Red Green, what's the difference between Type III sums
of squares and a trampoline? You take your shoes off to jump on a trampoline!
Type III sums of squares are the bane of ANOVA with unbalanced designs. At
least, you'd get that impression from listening to the R community. (See the
tutorial on unbalanced designs for details.) But sometimes you just got to
have them. Bill Venables, a staunch critic of Type III, let the cat out of
the box as to how to get them in R. Here is a function.
aovIII = function (formula, data = NULL)
{
options(contrasts = c("contr.sum", "contr.poly"))
aovIII.out <<- aov(formula, data)
print(drop1(aovIII.out, . ~ ., test = "F"))
options(contrasts = c("contr.treatment", "contr.poly"))
}
The syntax is the same as that of the aov()
function in so far as feeding it a formula and data are concerned, but there
is no need to save the output, as you're going to see it
all. Nevertheless, the function creates an aovIII.out object in your workspace,
which contains the output as if a Type I ANOVA had been calculated.
If you already have such an object there and don't wish it to be harmed,
CHANGE ITS NAME BEFORE RUNNING THIS FUNCTION!
> data(genotype, package="MASS") # get some unbalanced data
> summary(genotype) # weight of crossfostered mice by litter and mother
Litter Mother Wt
A:17 A:16 Min. :36.30
B:15 B:14 1st Qu.:48.20
I:14 I:16 Median :54.00
J:15 J:15 Mean :53.97
3rd Qu.:60.30
Max. :69.80
> with(genotype, table(Litter,Mother)) # check the cell frequencies
Mother
Litter A B I J
A 5 3 4 5
B 4 5 4 2
I 3 3 5 3
J 4 3 3 5
> aovIII(Wt ~ Mother * Litter, data=genotype) # the Type III tests
Single term deletions
Model:
Wt ~ Mother * Litter
Df Sum of Sq RSS AIC F value Pr(>F)
<none> 2440.8 257.04
Mother 3 671.74 3112.6 265.87 4.1282 0.01142 *
Litter 3 27.66 2468.5 251.73 0.1700 0.91612
Mother:Litter 9 824.07 3264.9 256.79 1.6881 0.12005
> summary(aovIII.out) # the Type I tests
Df Sum Sq Mean Sq F value Pr(>F)
Mother 3 771.6 257.20 4.742 0.00587 **
Litter 3 63.6 21.21 0.391 0.76000
Mother:Litter 9 824.1 91.56 1.688 0.12005
Residuals 45 2440.8 54.24
> rm(genotype, aov.outIII)
These are the data used by Bill Venables in his famous Exegeses,
in which he discusses unbalanced designs (among other things). One caution:
I've never really tested this method with repeated measures factors. I know it
works if all factors are between groups.
When I was teaching the senior research course here at CCU, I had all my
students put their data into a spreadsheet and send it to me, so I could use
R to check their statistical analysis. No matter how many times I instructed
them on the matter, I just couldn't get them to put repeated measures data into
a long-format data frame. So I wrote a function that would analyze the data in
short format (single factor designs only). Later, I expanded that function for
teaching purposes. Here is that function. (Don't ask me to explain it! I'm
not sure I remember!)
rmsaov.test = function(matname)
{
s <- dim(matname)[1]
df.subjects <- s - 1
a <- dim(matname)[2]
df.treatment <- a - 1
total.df <- a * s - 1
total.SS <- var(as.vector(matname)) * total.df
treatment.SS <- var(margin.table(matname, 2)) * df.treatment/s
treatment.MS <- treatment.SS/df.treatment
subjects.SS <- var(margin.table(matname, 1)) * df.subjects/a
subjects.MS <- subjects.SS/df.subjects
error.SS <- total.SS - treatment.SS - subjects.SS
df.error <- df.treatment * df.subjects
error.MS <- error.SS/df.error
STATISTIC <- treatment.MS/error.MS
PARAMETER <- c(df.treatment, df.error)
PVAL <- pf(STATISTIC, df.treatment, df.error, lower.tail = FALSE)
DNAME <- deparse(substitute(matname))
METHOD <- "Oneway ANOVA with repeated measures"
names(STATISTIC) <- "F"
names(PARAMETER) <- c("df treat", "df error")
OUT <- list(statistic = STATISTIC, parameter = PARAMETER,
p.value = PVAL, method = METHOD, data.name = DNAME, MS.treatment = treatment.MS,
MS.error = error.MS, MS.subjects = subjects.MS)
class(OUT) <- "htest"
print(OUT)
print(c(MS.error = error.MS, MS.treat = treatment.MS, MS.subjs = subjects.MS))
cat("\n")
cat("Treatment Means\n")
print(apply(matname, 2, mean))
cat("\n")
cat("Compound Symmetry Check (Variance-Covariance Matrix)\n")
covmat <- cov(matname)
print(covmat)
cat("\n")
cat("Estimated Greenhouse-Geisser Epsilon and Adjusted p-value\n")
D <- a^2 * (mean(diag(covmat)) - mean(covmat))^2
N1 <- sum(covmat^2)
N2 <- 2 * a * sum(apply(covmat, 1, mean)^2)
N3 <- a^2 * mean(covmat)^2
epsilon <- D/((a - 1) * (N1 - N2 + N3))
p.adj <- pf(treatment.MS/error.MS, df.treatment * epsilon,
df.error * epsilon, lower.tail = FALSE)
GGE <- c(epsilon, p.adj)
names(GGE) = c("epsilon", "p-adjust")
print(GGE)
}
As you can see, the function takes a single matrix as an argument. The matrix
should have the repeated measures data in columns (each time measurement or
each repeated measurement on the DV gets its own column). Each row of the
matrix corresponds to data from one subject.
Here are two examples of how to use this function. The first one involves
fetching some data from a help page for Friedman's test (a nonparametric
repeated measures ANOVA). The data are base running times for three different
methods of rounding first base. (The help page contains some errors, but see
for more details.)
> example(friedman.test) # output not shown; but there's a lot!
> ls()
[1] "aovIII" "rmsaov.test" "RoundingTimes" "wb"
> head(RoundingTimes) # the data matrix we are after
Round Out Narrow Angle Wide Angle
1 5.40 5.50 5.55
2 5.85 5.70 5.75
3 5.20 5.60 5.50
4 5.55 5.50 5.40
5 5.90 5.85 5.70
6 5.45 5.55 5.60
> class(RoundingTimes) # it must be a matrix
[1] "matrix"
> rmsaov.test(RoundingTimes)
Oneway ANOVA with repeated measures
data: RoundingTimes
F = 6.2883, df treat = 2, df error = 42, p-value = 0.004084
MS.error MS.treat MS.subjs
0.007451299 0.046856061 0.200887446
Treatment Means
Round Out Narrow Angle Wide Angle
5.543182 5.534091 5.459091
Compound Symmetry Check (Variance-Covariance Matrix)
Round Out Narrow Angle Wide Angle
Round Out 0.07387987 0.06310065 0.06327922
Narrow Angle 0.06310065 0.06747294 0.06705628
Wide Angle 0.06327922 0.06705628 0.07443723
Estimated Greenhouse-Geisser Epsilon and Adjusted p-value
epsilon p-adjust
0.773501463 0.008439799
The output is self-explanatory, but for more information see the tutorial on
repeated measures designs. The second example is from the anorexia data that
we have looked at before.
> rm(RoundingTimes, wb) # clean up a bit first
> data(anorexia, package="MASS") # fetch the data
> head(anorexia)
Treat Prewt Postwt
1 Cont 80.7 80.2
2 Cont 89.4 80.1
3 Cont 91.8 86.4
4 Cont 74.0 86.3
5 Cont 78.1 76.1
6 Cont 88.3 78.1
> class(anorexia) # this is no good!
[1] "data.frame"
> anor = as.matrix(anorexia[,2:3]) # convert the relevant columns to a matrix
> rmsaov.test(anor)
Oneway ANOVA with repeated measures
data: anor
F = 8.6293, df treat = 1, df error = 71, p-value = 0.004458
MS.error MS.treat MS.subjs
31.86892 275.00694 59.55305
Treatment Means
Prewt Postwt
82.40833 85.17222
Compound Symmetry Check (Variance-Covariance Matrix)
Prewt Postwt
Prewt 26.85796 13.84207
Postwt 13.84207 64.56401
Estimated Greenhouse-Geisser Epsilon and Adjusted p-value
epsilon p-adjust
1.000000000 0.004457718
That's it for ANOVA-related functions. Now we must enhance our functions file
with the two new ones we've created.
> ls()
[1] "anor" "anorexia" "aovIII" "rmsaov.test"
> rm(anor, anorexia) # get rid of the junk
> ls() # check!
[1] "aovIII" "rmsaov.test"
> load("myfunctions.RData") # add the functions in myfunctions
> ls() # check again!
[1] "aovIII" "bygroups" "Cohens.d" "error.bars" "r.crit" "rmsaov.test"
[7] "sampsize" "sem" "SS" "summarize"
> save.image("myfunctions.RData") # save a new myfunctions file
> rm(list=ls()) # clean up
> ls()
character(0)
One more new function and we'll call it a day.
Linear Contrasts
There is no easy way in the base R packages to test complex linear contrasts.
So we're going to create one. The method of calculation will be based on that in
the very excellent statistics book The Statistical Sleuth (3rd ed.) by Ramsey
and Schafer (2013).
t.contrast = function(means,codes,n,spool,dfe)
{
g = sum(means*codes)
seg = spool*sqrt(sum(codes^2/n))
t = g/seg
p.value = pt(abs(t),dfe,lower.tail=FALSE)*2
d = abs(g/spool)
output <<- c(g=g,SE=seg,t=t,df=dfe,p.value=p.value,d=d)
round(output,4)
}
Executing this function will write an object to your workspace called "output",
so be careful about that. If you don't want that to happen, modify the script
so that the line that begins output <<- is output <- (a single arrowhead on
the assignment arrow).
This is how the function works. Linear contrasts are used as an alternative
to ANOVA and are sometimes referred to as planned comparisons. ANOVA is a fine
tool, but it tests an alternative hypothesis that is very general, i.e., any
pattern of means other than all means equal. Thus, ANOVA is most appropriate
when you don't really know much about what to expect from your treatments. If
you have specific hypotheses, for example, group A will have a higher mean
than group B, and groups B and C will be the same, then planned comparisons are
more appropriate.
> ls() # your workspace should contain only the function
[1] "t.contrast"
> data(chickwts) # weights of chickens by type of feed they received
> summary(chickwts)
weight feed
Min. :108.0 casein :12
1st Qu.:204.5 horsebean:10
Median :258.0 linseed :12
Mean :261.3 meatmeal :11
3rd Qu.:323.5 soybean :14
Max. :423.0 sunflower:12
I don't really know much about feeding chickens other than that I had to do it
as a kid and didn't care for it. Here we have 71 chickens in six groups by the
type of feed they are receiving. The DV is weight. Let's pretend we (I) know
enough about chicken feed to have the hypothesis that casein and sunflower are
the best feeds, and are about equally good. The other four are not as good and
are about equally not as good. So we're expecting the casein and sunflower means
to be high and roughly equal, and the other four means to be low and roughly
equal. In contrast codes (I'll have to refer you to an outside source here),
that hypothesis would be expressed as follows: (1/2, -1/4, -1/4, -1/4, -1/4, 1/2).
That will have the effect of combining the first and last group into one big
"megagroup" and the middle four groups into another "megagroup," and then
those two "megagroups" will be tested against each other.
We need to feed our t.contrast function five pieces of information: a vector
of group means, a vector of contrast codes, a vector of group sizes (can be one
number if the design is balanced), the pooled standard deviation calculated over
all groups, and the degrees of freedom for error, which is total number of
subjects minus the number of groups they've been divided into if the design is
between groups. Let's start getting that stuff.
> means = with(chickwts, tapply(weight,feed,mean))
> codes = c(1/2, -1/4, -1/4, -1/4, -1/4, 1/2) # best entered as fractions
> n = c(12, 10, 12, 11, 14, 12) # from the summary above
> aov(weight~feed, data=chickwts) # easiest way to get spool
Call:
aov(formula = weight ~ feed, data = chickwts)
Terms:
feed Residuals
Sum of Squares 231129.2 195556.0
Deg. of Freedom 5 65
Residual standard error: 54.85029
> spool = 54.85029 # it's the residual s.e.
> dfe = 65
And now we're ready to run the function.
> t.contrast(means, codes, n, spool, dfe)
g SE t df p.value d
100.6781 13.7969 7.2972 65.0000 0.0000 1.8355
g is the difference in means between the two "megagroups," SE is the standard
error (estimated) of that difference, t is the corresponding test statistic,
df and p.value should be obvious, and d is Cohen's d for the difference g.
We could (and probably should) continue with further contrasts, but I'll
leave that to you as an exercise (as authors always say in these sorts of
situations when they really don't want to do something).
Now let's add the function to our file. First, make sure there is nothing
in your workspace except this function.
> ls()
[1] "chickwts" "codes" "dfe" "means" "n"
[6] "output" "spool" "t.contrast"
> rm(chickwts, codes, dfe, means, n, output, spool)
> ls()
[1] "t.contrast"
And we made quite a mess while testing the function, so we had to clean it up.
Otherwise, all that junk would be going into the myfunctions file.
> load("myfunctions.RData")
> ls()
[1] "aovIII" "bygroups" "Cohens.d" "error.bars" "r.crit" "rmsaov.test"
[7] "sampsize" "sem" "SS" "summarize" "t.contrast"
> save.image("myfunctions.RData")
> rm(list=ls())
There is one more thing I want to do to prepare us for the next section of
this tutorial.
> data(anorexia, package="MASS") # we'll use this below
> save.image("anorexia.RData") # a saved project
> rm(anorexia) # clean up
> palm483S = rnorm(50) # some junk for the workspace
We are ready to move on.
Putting It All Together and Using It
We now have eleven shiny new functions in a file called myfunctions.RData. In
my case, I have this file saved to my Rspace directory, because that's usually
where I go to do my R work. You should adjust the advice here to fit your own
working habits.
When I start R, I immediately change my working directory to Rspace. Then I
examine my workspace, and if there is stuff there I don't want, I remove it.
I load anything from Rspace that I might be working on. Then I'm ready to go
to work.
Suppose I wanted to use one or more of the functions we've just created.
It's an RData file, so I could load() it, but
that would put all those functions in my workspace, and I don't want that.
So instead of loading it, I'm going to attach it to the search path. My
work flow might look like this.
> setwd("Rspace") # don't do this if you're already in Rspace
> ls()
[1] "palm483S"
> rm(palm483S)
> attach("myfunctions.RData")
Those functions are now in my search path, and if I call one of them, R will
be able to find it.
> search()
[1] ".GlobalEnv" "file:myfunctions.RData" "tools:RGUI"
[4] "package:stats" "package:graphics" "package:grDevices"
[7] "package:utils" "package:datasets" "package:methods"
[10] "Autoloads" "package:base"
> ls(pos=2)
[1] "aovIII" "bygroups" "Cohens.d" "error.bars" "r.crit" "rmsaov.test"
[7] "sampsize" "sem" "SS" "summarize" "t.contrast"
Just in case you didn't believe me!
> dir() # see what's in Rspace
[1] "anorexia.RData" "myfunctions.RData" "UCB.RData"
> load("anorexia.RData") # load the previously saved project I want
> ls() # nobody here but us data sets
[1] "anorexia"
> head(anorexia)
Treat Prewt Postwt
1 Cont 80.7 80.2
2 Cont 89.4 80.1
3 Cont 91.8 86.4
4 Cont 74.0 86.3
5 Cont 78.1 76.1
6 Cont 88.3 78.1
> dim(anorexia)
[1] 72 3
> with(anorexia, cor(Prewt, Postwt)) # do some analysis
[1] 0.3324062
> r.crit(df=70) # use one of our custom functions
df alpha 1-tail 2-tail
70.0000 0.0500 0.1954 0.2319
> bygroups(data=anorexia$Postwt, groups=anorexia$Treat) # use another one
CBT Cont FT
85.69655 81.10769 90.49412
> summarize(anorexia$Prewt) # caught me again; this is the old one
mean SD N
82.408333 5.182466 72.000000
> detach("file:myfunctions.RData") # done with it; get it out of the search path
It's as easy as that!
Review of Steps For Modifying Your Functions File
You may want to add a function of your own to this file, or delete one. Or
you may want to modify one of these functions. Just to collect the steps
together in a neat outline, here's how you would do it. Make sure the file
is not attached to your search path before you begin.
To delete a function (or a data object that snuck in there somehow):
- Change to the working directory that has the myfunctions.RData file in it.
- Clear your workspace.
- load("myfunctions.RData")
- Remove the undesired function or functions from your workspace using
rm().
- save.image("myfunctions.RData")
- Clear your workspace.
To add a function:
- Change to the working directory that has the myfunctions.RData file in it.
- Clear your workspace.
- Write the new function in the script editor (or in a text editor) and
read it into R (Execute or Run All from the script window, or copy and paste).
I.e., get it into your workspace, and make sure it's the ONLY thing in your
workspace.
- Make sure the new function doesn't have the same name as an old one, or it
will be overwritten at the next step.
- load("myfunctions.RData")
- save.image("myfunctions.RData")
- Clear your workspace.
To modify a function:
- Change to the working directory that has the myfunctions.RData file in it.
- Clear your workspace.
- load("myfunctions.RData")
- Type the name of the function you wish to modify at the command prompt
without parentheses. This will print the definition of the function to your
Console. Example:
> aovIII
function (formula, data = NULL, subset=NULL)
{
options(contrasts = c("contr.sum", "contr.poly"))
aov.out <<- aov(formula, data, subset)
print(drop1(aov.out, . ~ ., test = "F"))
options(contrasts = c("contr.treatment", "contr.poly"))
}
- Copy and paste the function definition to your script window and insert
the function name and an = sign before the word function. (NOTE: You can also
try fix(aovIII), which may or may not work. In
this case, DO NOT type in the function name =.)
- Make the desired mods. For example, if you don't want aovIII to save the
aov.out object to your workspace, modify the aov.out <<- line to aov.out <-.
- Read the modified function into R (or close the window opened by fix and
choose Save).
- save.image("myfunctions.RData")
- Clear your workspace.
I should also note that you can add other objects, such as data frames,
vectors, matrices, etc., to this file if you want to keep them there for
some reason. I have a long list of data frames in mine that I use in my
teaching. Sending the file to my students by e-mail is a convenient way to
get a large number of data files to them. (No, I won't send it to you.
Some of the data files were collected by students and are not for public
use. Sorry.)
An Alternative Method
If you don't really need all your scripts and data files in one big file
such as myfunctions.RData, then it may be more convenient to just save them a
script at a time in your working directory (Rspace). Open a script window,
type the script, then save it with a convenient name and .R as the extension.
Be sure R is saving it to your working directory. This is not the default in
the script editor, at least not on the Mac, so make the necessary changes in
the dialogue box before saving.
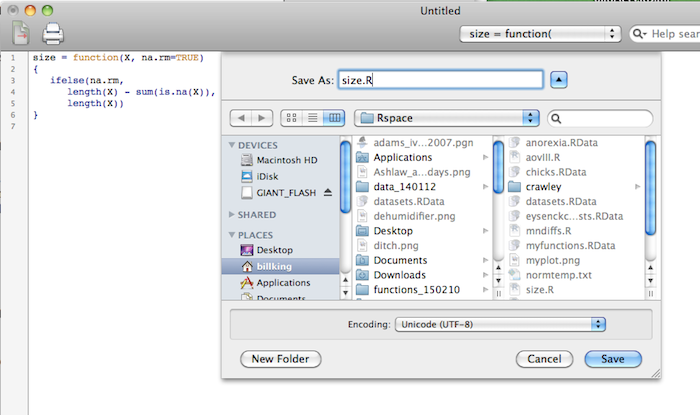
Then when you need to use it, you just source it in.
> setwd("Rspace")
> source("size.R")
> size(c(NA,1:5,NA,NA,NA))
[1] 5
If you need to modify the script at some point, you don't need to go
through all the gyrations above. Just open the script in a script window,
make whatever changes you want, and save it. The disadvantage is, if you
have a large number of scripts, you might end up making a mess of your
working directory. Then again, you could always create a subdirectory. Call
it, oh, say, "functions".
created 2016 January 24; modified 2016 April 7
| Table of Contents
| Function Reference
| Function Finder
| R Project |
|

