
| Table of Contents
| Function Reference
| Function Finder
| R Project |
LOGISTIC REGRESSION
Preliminaries
Model Formulae
You will need to know a bit about Model Formulae
to understand this tutorial.
Odds, Odds Ratios, and Logit
When you go to the track, how do you know which horse to bet on? You look at
the odds. In the program, you may see the odds for your horse, Sea Brisket, are
8 to 1, which are the odds AGAINST winning. This means in nine races Sea Brisket
would be expected to win 1 and lose 8. In probability terms, Sea Brisket has a
probability of winning of 1/9, or 0.111. But the odds of winning are 1:8, 1/8, or
0.125. Odds are actually the ratio of two probabilities.
p(one outcome) p(success) p
odds = -------------------- = ----------- = ---, where q = 1 - p
p(the other outcome) p(failure) q
So for Sea Brisket, odds(winning) = (1/9)/(8/9) = 1/8. Notice that odds have
these properties:
- If p(success) = p(failure), then odds(success) = 1 (or 1 to 1, or 1:1).
- If p(success) < p(failure), then odds(success) < 1.
- If p(success) > p(failure), then odds(success) > 1.
- Unlike probability, which cannot exceed 1, there is no upper bound on odds.
The natural log of odds is called the logit, or logit transformation, of p:
logit(p) = loge(p/q). Logit is sometimes called "log
odds." Because of the properties of odds given in the list above, the logit has
these properties:
- If odds(success) = 1, then logit(p) = 0.
- If odds(success) < 1, then logit(p) < 0.
- If odds(success) > 1, then logit(p) > 0.
- The logit transform fails if p = 0.
Logistic regression is a method for fitting a regression curve,
y = f(x), when y consists of proportions or probabilities, or binary
coded (0,1--failure, success) data. When the response is a binary
(dichotomous)
variable, and x is numeric, logistic regression fits a logistic curve to the
relationship between x and y. The logistic curve looks like the graph at
right. It is an S-shaped or sigmoid curve, often used to model population
growth, survival from a disease, the spread of a disease (or something
disease-like, such as a rumor), and so on. The logistic function is...
y = [exp(b0 + b1x)] / [1 + exp(b0 + b1x)]
Logistic regression fits b0 and b1, the regression
coefficients (which were 0 and 1, respectively, for the graph above). It should
have already struck you that this curve is not linear. However, the point of the
logit transform is to make it linear.
logit(y) = b0 + b1x
Hence, logistic regression is linear regression on the logit transform of y,
where y is the proportion (or probability) of success at each value of x.
However, you should avoid the temptation to do a traditional least-squares
regression at this point, as neither the normality nor the homoscedasticity
assumption will be met.
The odds ratio might best be illustrated by returning to our horse race.
Suppose in the same race, Seattle Stew is given odds of 2 to 1 against, which
is to say,
two expected loses for each expected win. Seattle Stew's odds of winning are
1/2, or 0.5. How much better is this than the winning odds for Sea Brisket? The
odds ratio tells us: 0.5 / 0.125 = 4.0. The odds of Seattle
Stew winning are four times the odds of Sea Brisket winning. Be careful not to
say "times as likely to win," which would not be correct. The probability
(likelihood, chance) of Seattle Stew winning is 1/3 and for Sea Brisket is 1/9,
resulting in a likelihood ratio of 3.0. Seattle Stew is three times more
likely to win than is Sea Brisket.
Logistic Regression: One Numeric Predictor
In the "MASS" library there is a data set called "menarche" (Milicer, H. and
Szczotka, F., 1966, Age at Menarche in Warsaw girls in 1965, Human Biology, 38,
199-203), in which there are three variables: "Age" (average age of age
homogeneous groups of girls), "Total" (number of girls in each group), and
"Menarche" (number of girls in the group who have reached menarche).
> library("MASS")
> data(menarche)
> str(menarche)
'data.frame': 25 obs. of 3 variables:
$ Age : num 9.21 10.21 10.58 10.83 11.08 ...
$ Total : num 376 200 93 120 90 88 105 111 100 93 ...
$ Menarche: num 0 0 0 2 2 5 10 17 16 29 ...
> summary(menarche)
Age Total Menarche
Min. : 9.21 Min. : 88.0 Min. : 0.00
1st Qu.:11.58 1st Qu.: 98.0 1st Qu.: 10.00
Median :13.08 Median : 105.0 Median : 51.00
Mean :13.10 Mean : 156.7 Mean : 92.32
3rd Qu.:14.58 3rd Qu.: 117.0 3rd Qu.: 92.00
Max. :17.58 Max. :1049.0 Max. :1049.00
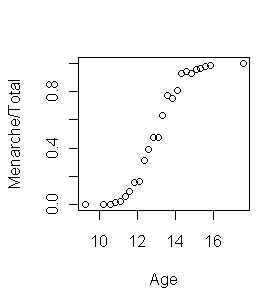
> plot(Menarche/Total ~ Age, data=menarche)
From the graph at right, it appears a logistic fit is called for here. The fit
would be done this way.
> glm.out = glm(cbind(Menarche, Total-Menarche) ~ Age, family=binomial(logit), data=menarche)
Numerous explanation are in order! First, glm()
is the function used to do generalized linear models. With "family=" set to
"binomial" with a "logit" link, glm()
produces a logistic regression. Because we are using
glm() with binomial errors in the response variable, the ordinary
assumptions of least squares linear regression (normality and homoscedasticity)
don't apply. Second, our data frame does not contain a row for every case
(i.e., every girl upon whom data were collected). Therefore, we do not have a
binary (0,1) coded response variable. No problem! If we feed glm() a table (or matrix) in which the
first column is number of successes and the second column is number of
failures, R will take care of the coding for us. In the above analysis, we made
that table on the fly inside the model formula by binding "Menarche" and
"Total − Menarche" into the columns of a table (matrix) using
cbind().
Let's look at how closely the fitted values from our logistic regression
match the observed values.
> plot(Menarche/Total ~ Age, data=menarche)
> lines(menarche$Age, glm.out$fitted, type="l", col="red")
> title(main="Menarche Data with Fitted Logistic Regression Line")
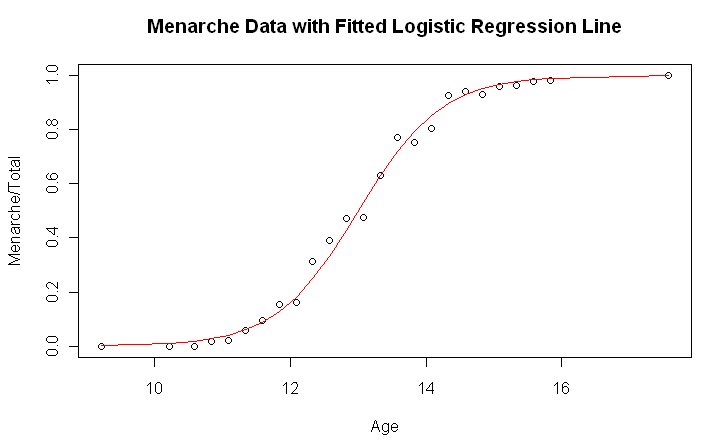 I'm impressed! The numerical results are extracted like this.
I'm impressed! The numerical results are extracted like this.
> summary(glm.out)
Call:
glm(formula = cbind(Menarche, Total - Menarche) ~ Age, family = binomial(logit),
data = menarche)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.0363 -0.9953 -0.4900 0.7780 1.3675
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -21.22639 0.77068 -27.54 <2e-16 ***
Age 1.63197 0.05895 27.68 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 3693.884 on 24 degrees of freedom
Residual deviance: 26.703 on 23 degrees of freedom
AIC: 114.76
Number of Fisher Scoring iterations: 4
The following requests also produce useful results: glm.out$coef,
glm.out$fitted, glm.out$resid, glm.out$effects, and anova(glm.out).
Recall that the response variable is log odds, so the coefficient of "Age"
can be interpreted as "for every one year increase in age the odds of having
reached menarche increase by exp(1.632) = 5.11 times."
To evaluate the overall performance of the model, look at the null deviance
and residual deviance near the bottom of the print out. Null deviance shows how
well the response is predicted by a model with nothing but an intercept (grand
mean). This is essentially a chi square value on 24 degrees of freedom, and
indicates very little fit (a highly significant difference between fitted values
and observed values). Adding in our predictors--just "Age" in this case--decreased
the deviance by 3667 points on 1 degree of freedom. Again, this is interpreted
as a chi square value and indicates a highly significant decrease in deviance.
The residual deviance is 26.7 on 23 degrees of freedom. We use this to test the
overall fit of the model by once again treating this as a chi square value. A
chi square of 26.7 on 23 degrees of freedom yields a p-value of 0.269. The null
hypothesis (i.e., the model) is not rejected. The fitted values are not
significantly different from the observed values.
Logistic Regression: Multiple Numerical Predictors
During the Fall semester of 2005, two students in our program--Rachel Mullet
and Lauren Garafola--did a senior research project in which they studied a
phenomenon called Inattentional Blindness (IB). IB refers to situations in which
a person fails to see an obvious stimulus right in front of his eyes. (For
details see this website.)
In their study, Rachel and Lauren had subjects view an online video showing
college students passing basketballs to each other, and the task was to count
the number of times students in white shirts passed the basketball. During the
video, a person in a black gorilla suit walked though the picture in a very
obvious way. At the end of the video, subjects were asked if they saw the
gorilla. Most did not!
Rachel and Lauren hypothesized that IB could be predicted from performance
on the Stroop Color Word test. This test produces three scores: "W" (word alone,
i.e., a score derived from reading a list of color words such as red, green,
black), "C" (color alone, in which a score is derived from naming the color in
which a series of Xs are printed), and "CW" (the Stroop task, in which a score
is derived from the subject's attempt to name the color in which a color word
is printed when the word and the color do not agree). The data are in the
following table, in which the response, "seen", is coded as 0=no and 1=yes.
gorilla = read.table(header=T, text="
seen W C CW
0 126 86 64
0 118 76 54
0 61 66 44
0 69 48 32
0 57 59 42
0 78 64 53
0 114 61 41
0 81 85 47
0 73 57 33
0 93 50 45
0 116 92 49
0 156 70 45
0 90 66 48
0 120 73 49
0 99 68 44
0 113 110 47
0 103 78 52
0 123 61 28
0 86 65 42
0 99 77 51
0 102 77 54
0 120 74 53
0 128 100 56
0 100 89 56
0 95 61 37
0 80 55 36
0 98 92 51
0 111 90 52
0 101 85 45
0 102 78 51
1 100 66 48
1 112 78 55
1 82 84 37
1 72 63 46
1 72 65 47
1 89 71 49
1 108 46 29
1 88 70 49
1 116 83 67
1 100 69 39
1 99 70 43
1 93 63 36
1 100 93 62
1 110 76 56
1 100 83 36
1 106 71 49
1 115 112 66
1 120 87 54
1 97 82 41
")
We might begin like this.
> cor(gorilla) # a correlation matrix
seen W C CW
seen 1.00000000 -0.03922667 0.05437115 0.06300865
W -0.03922667 1.00000000 0.43044418 0.35943580
C 0.05437115 0.43044418 1.00000000 0.64463361
CW 0.06300865 0.35943580 0.64463361 1.00000000
Or like this.
> with(gorilla, tapply(W, seen, mean))
0 1
100.40000 98.89474
> with(gorilla, tapply(C, seen, mean))
0 1
73.76667 75.36842
> with(gorilla, tapply(CW, seen, mean))
0 1
46.70000 47.84211
The Stroop scale scores are moderately positively correlated with each other,
but none of them appears to be related to the "seen" response variable, at
least not to any impressive extent. There doesn't appear to be much here to
look at. Let's have a go at it anyway. Since the response is a binomial
variable, a logistic regression can be done as follows.
> glm.out = glm(seen ~ W * C * CW, family=binomial(logit), data=gorilla)
> summary(glm.out)
Call:
glm(formula = seen ~ W * C * CW, family = binomial(logit), data = gorilla)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.8073 -0.9897 -0.5740 1.2368 1.7362
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -1.323e+02 8.037e+01 -1.646 0.0998 .
W 1.316e+00 7.514e-01 1.751 0.0799 .
C 2.129e+00 1.215e+00 1.753 0.0797 .
CW 2.206e+00 1.659e+00 1.329 0.1837
W:C -2.128e-02 1.140e-02 -1.866 0.0621 .
W:CW -2.201e-02 1.530e-02 -1.439 0.1502
C:CW -3.582e-02 2.413e-02 -1.485 0.1376
W:C:CW 3.579e-04 2.225e-04 1.608 0.1078
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 65.438 on 48 degrees of freedom
Residual deviance: 57.281 on 41 degrees of freedom
AIC: 73.281
Number of Fisher Scoring iterations: 5
> anova(glm.out, test="Chisq")
Analysis of Deviance Table
Model: binomial, link: logit
Response: seen
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev P(>|Chi|)
NULL 48 65.438
W 1 0.075 47 65.362 0.784
C 1 0.310 46 65.052 0.578
CW 1 0.106 45 64.946 0.745
W:C 1 2.363 44 62.583 0.124
W:CW 1 0.568 43 62.015 0.451
C:CW 1 1.429 42 60.586 0.232
W:C:CW 1 3.305 41 57.281 0.069
Two different extractor functions have been used to see the results of our
analysis. What do they mean?
The first gives us what amount to regression coefficients with standard
errors and a z-test, as we saw in the single variable example above. None of the
coefficients are significantly different from zero (but a few are close). The
deviance was reduced by 8.157 points on 7 degrees of freedom, for a p-value
of...
> 1 - pchisq(8.157, df=7)
[1] 0.3189537
Overall, the model appears to have performed poorly, showing no significant
reduction in deviance (no significant difference from the null model).
The second print out shows the same overall reduction in deviance, from
65.438 to 57.281 on 7 degrees of freedom. In this print out, however, the
reduction in deviance is shown for each term, added sequentially first to last.
Of note is the three-way interaction term, which produced a nearly significant
reduction in deviance of 3.305 on 1 degree of freedom (p = 0.069).
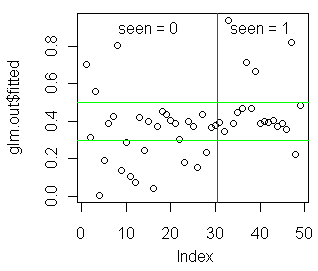
In the event you are encouraged by any of this, the following graph might be
revealing.
> plot(glm.out$fitted)
> abline(v=30.5,col="red")
> abline(h=.3,col="green")
> abline(h=.5,col="green")
> text(15,.9,"seen = 0")
> text(40,.9,"seen = 1")
I'll leave it up to you to interpret this on your own time. (Also, try
running the anova() tests on a model in which CW
has been entered first.)
Logistic Regression: Categorical Predictors
Let's re-examine the "UCBAdmissions" data set, which we looked at in a
previous tutorial.
> ftable(UCBAdmissions, col.vars="Admit")
Admit Admitted Rejected
Gender Dept
Male A 512 313
B 353 207
C 120 205
D 138 279
E 53 138
F 22 351
Female A 89 19
B 17 8
C 202 391
D 131 244
E 94 299
F 24 317
The data are from 1973 and show admissions by gender to the top six grad
programs at the University of California, Berkeley. Looked at as a two-way
table, there appears to be a bias against admitting women.
> dimnames(UCBAdmissions)
$Admit
[1] "Admitted" "Rejected"
$Gender
[1] "Male" "Female"
$Dept
[1] "A" "B" "C" "D" "E" "F"
> margin.table(UCBAdmissions, c(2,1))
Admit
Gender Admitted Rejected
Male 1198 1493
Female 557 1278
However, there are also relationships between "Gender" and "Dept" as well as
between "Dept" and "Admit", which means the above relationship may be confounded
by "Dept" (or "Dept" might be a lurking variable, in the language of traditional
regression analysis). Perhaps a logistic regression with the binomial variable
"Admit" as the response can tease these variables apart.
If there is a way to conveniently get that flat table into a data frame
(without splitting an infinitive), I don't know it. So I had to do this. (I'll
leave off the command prompts so you can copy and paste it.)
### begin copying here
ucb.df = data.frame(gender=rep(c("Male","Female"),c(6,6)),
dept=rep(LETTERS[1:6],2),
yes=c(512,353,120,138,53,22,89,17,202,131,94,24),
no=c(313,207,205,279,138,351,19,8,391,244,299,317))
### end copying here and paste into the R Console
> ucb.df
gender dept yes no
1 Male A 512 313
2 Male B 353 207
3 Male C 120 205
4 Male D 138 279
5 Male E 53 138
6 Male F 22 351
7 Female A 89 19
8 Female B 17 8
9 Female C 202 391
10 Female D 131 244
11 Female E 94 299
12 Female F 24 317
Once again, we do not have a binary coded response variable, so the last two
columns of this data frame will have to be bound into the columns of a table
(matrix) to serve as the response in the model formula.
> mod.form = "cbind(yes,no) ~ gender * dept" # mind the quotes here!
> glm.out = glm(mod.form, family=binomial(logit), data=ucb.df)
I used a trick here of storing the model formula in a data object, and then
entering the name of this object into the
glm() function. That way, if I
made a mistake in the model formula (or want to run an alternative model), I
have only to edit the "mod.form" object to do it.
Let's see what we have found.
> options(show.signif.stars=F) # turn off significance stars (optional)
> anova(glm.out, test="Chisq")
Analysis of Deviance Table
Model: binomial, link: logit
Response: cbind(yes, no)
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev P(>|Chi|)
NULL 11 877.06
gender 1 93.45 10 783.61 4.167e-22
dept 5 763.40 5 20.20 9.547e-163
gender:dept 5 20.20 0 -2.265e-14 1.144e-03
This is a saturated model, meaning we have used up all our degrees of freedom,
and there is no residual deviance left over at the end. Saturated models always
fit the data perfectly. In this case, it appears the saturated model is required
to explain the data adequately. If we leave off the interaction term, for
example, we will be left with a residual deviance of 20.2 on 5 degrees of
freedom, and the model will be rejected (p = .001144). It appears all
three terms are making a significant contribution to the model.
How they are contributing appears if we use the other extractor.
> summary(glm.out)
Call:
glm(formula = mod.form, family = binomial(logit), data = ucb.df)
Deviance Residuals:
[1] 0 0 0 0 0 0 0 0 0 0 0 0
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.5442 0.2527 6.110 9.94e-10
genderMale -1.0521 0.2627 -4.005 6.21e-05
deptB -0.7904 0.4977 -1.588 0.11224
deptC -2.2046 0.2672 -8.252 < 2e-16
deptD -2.1662 0.2750 -7.878 3.32e-15
deptE -2.7013 0.2790 -9.682 < 2e-16
deptF -4.1250 0.3297 -12.512 < 2e-16
genderMale:deptB 0.8321 0.5104 1.630 0.10306
genderMale:deptC 1.1770 0.2996 3.929 8.53e-05
genderMale:deptD 0.9701 0.3026 3.206 0.00135
genderMale:deptE 1.2523 0.3303 3.791 0.00015
genderMale:deptF 0.8632 0.4027 2.144 0.03206
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 8.7706e+02 on 11 degrees of freedom
Residual deviance: -2.2649e-14 on 0 degrees of freedom
AIC: 92.94
Number of Fisher Scoring iterations: 3
These are the regression coefficients for each predictor in the model, with the
base level of each factor being suppressed. Remember, we are predicting log
odds, so to make sense of these coefficients, they need to be "antilogged".
> exp(-1.0521) # antilog of the genderMale coefficient
[1] 0.3492037
> 1/exp(-1.0521)
[1] 2.863658
This shows that men were actually at a significant disadvantage when
department and the interaction are controlled. The odds of a male being admitted
were only 0.35 times the odds of a female being admitted. The reciprocal of this
turns it on its head. All else being equal, the odds of female being admitted
were 2.86 times the odds of a male being admitted. (Any excitement we may be
experiencing over these "main effects" should be tempered by the fact that we
have a significant interaction that needs interpreting.)
Each coefficient compares the corresponding predictor to the base level. So...
> exp(-2.2046)
[1] 0.1102946
...the odds of being admitted to department C were only about 1/9th the odds of
being admitted to department A, all else being equal. If you want to compare,
for example, department C to department D, do this.
> exp(-2.2046) / exp(-2.1662) # C:A / D:A leaves C:D
[1] 0.962328
All else equal, the odds of being admitted to department C were 0.96 times the
odds of being admitted to department D.
To be honest, I'm not sure I'm comfortable with the interaction in this
model. It seems to me the last few calculations we've been doing would make more
sense without it. You might want to examine the interaction, and if you think it
doesn't merit inclusion, run the model again without it. Try doing that entering
"dept" first.
> mod.form="cbind(yes,no) ~ dept + gender"
> glm.out=glm(mod.form, family=binomial(logit), data=ucb.df)
> anova(glm.out, test="Chisq")
Statistics are nice, but in the end it's what makes sense that
should rule the day.
revised 2016 February 19
| Table of Contents
| Function Reference
| Function Finder
| R Project |
|




